In addition to LC3 protein, p62 or other linker molecules can also be used as markers of autophagy. Based on the characteristic of autophagy adaptor protein degradation through a selective autophagy pathway, Creative Bioarray can provide you with accurate and efficient autophagy detection.
In addition to LC3, levels of p62 (also known as SQSTM1/sequestosome 1) can also be used to monitor autophagic flux. p62 possesses a C-terminal ubiquitin-binding domain and a short LC3-interacting region sequence. It serves as an adaptor protein that links ubiquitinated proteins to the autophagic machinery and enables their clearance in the lysosome. p62 and p62-bound ubiquitinated proteins become incorporated into the completed autophagosome and are degraded in autolysosomes. Since the intracellular content of such adaptor molecules is obviously regulated by the level of autophagy, the protein level of adaptor molecules in the cell can reflect the autophagy flow in the cell. P62 is mainly removed by autophagy, and its amount is generally considered to be inversely proportional to autophagy activity.
There are many methods for studying autophagy based on the level of P62/SQSTM1. Creative Bioarray focuses on technological improvement and innovation, and can provide you with more accurate and efficient research services. Our services include but are not limited to:
P62/SQSTM1-luciferase Testing Service
Creative Bioarray has improved the P62/SQSTM1-based detection method by constructing a cell line stably expressing SQSTM1-luc protein, which is more suitable for throughput screening.
We constructed a recombinant plasmid expressing the selective autophagy substrate p62/SQSTM1 (Sequistosome1) and the firefly luciferase reporter gene. By detecting the activity of intracellular luciferase, autophagy flow can be analyzed.
Experiment Process
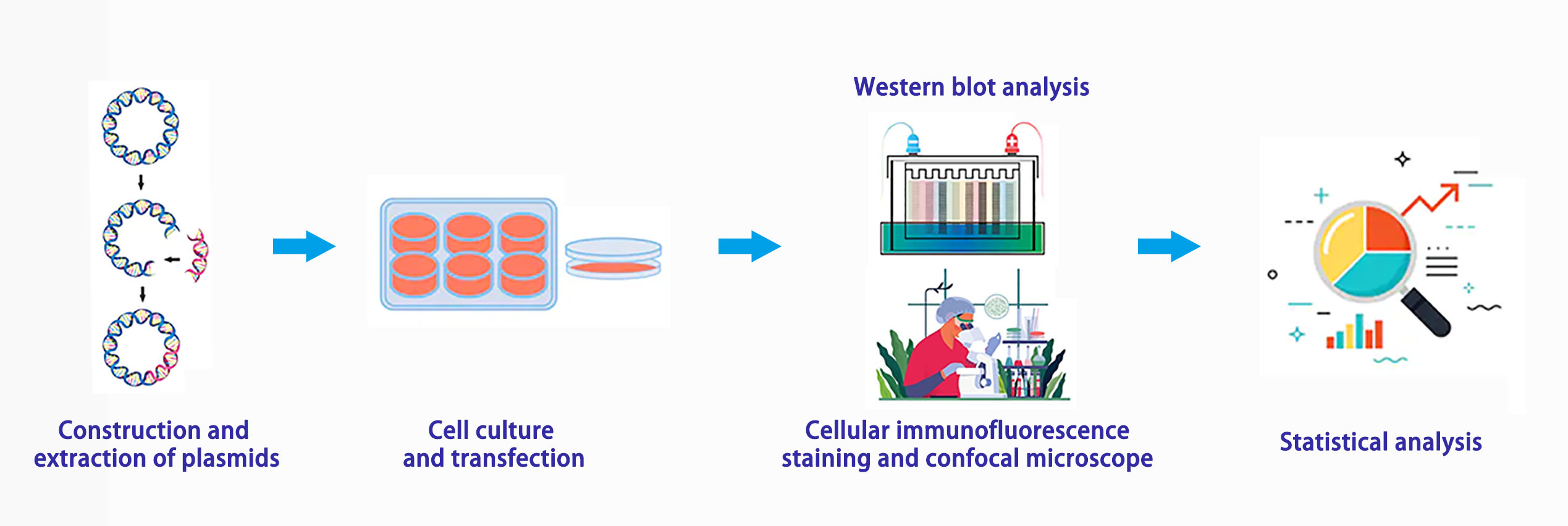
Advantages
The time depends on the experiment content
If you are interested in our services, please contact us for more detailed information.
Online Inquiry